Table of contents
MutRank: an R shiny web-application for exploratory targeted mutual rank-based coexpression analyses
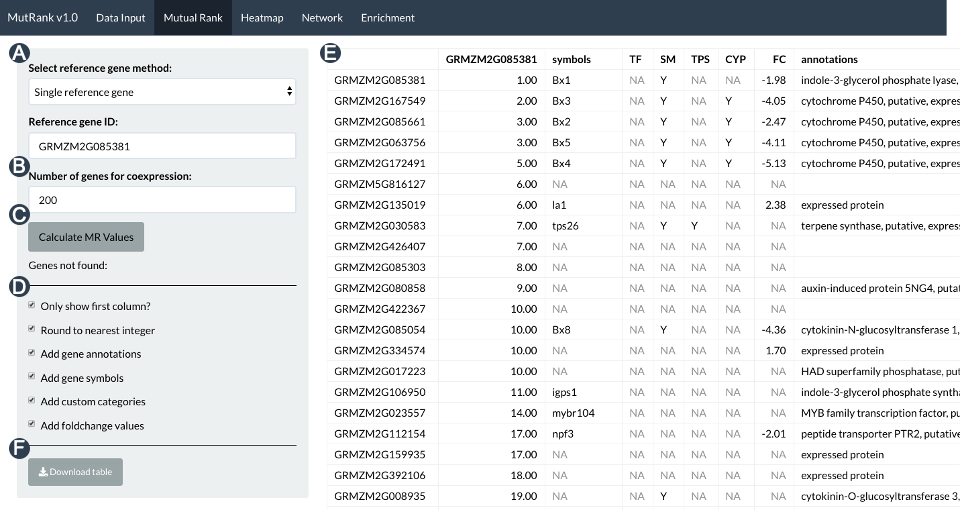
Transcriptomic data can uncover complex biological processes in part through the improved understanding of gene coexpression patterns. Mutual Rank (MR), the geometric mean of the ranked Pearson’s Correlation Coefficient (PCCs) between a pair of genes was shown to be a better indicator of functional associations and produces more robust results when using raw data compared to PCC. MR analyses were shown to be highly effective for identifying highly correlated specialized metabolism pathway genes, serving as springboard for hypothesis testing and validation. I developed MutRank web-app using the R Shiny package, to facilitate user control over both targeted and non-targeted MR-based coexpression analyses for rapid hypothesis testing. In addition to identifying highly coexpressed genes in any user-provided expression dataset, MutRank automatically integrates supporting information such as gene annotations, differential-expression data, predicted domains and assigned GO terms and provides useful tabular and graphical outputs as foundation for empirical hypothesis testing. The goal of MutRank is to provide simple, customizable and readily accessible tools to speed research progress in connecting metabolic phenotypes to genotypes for the purpose of understanding biological roles. More detailed information are available on the MutRank GitHub page and manuscript, linked above.
Data input menu 
Mutual rank menu