I have been working on creating Colab notebooks for a number of bioinformatics projects that I have been working on.
Table of contents
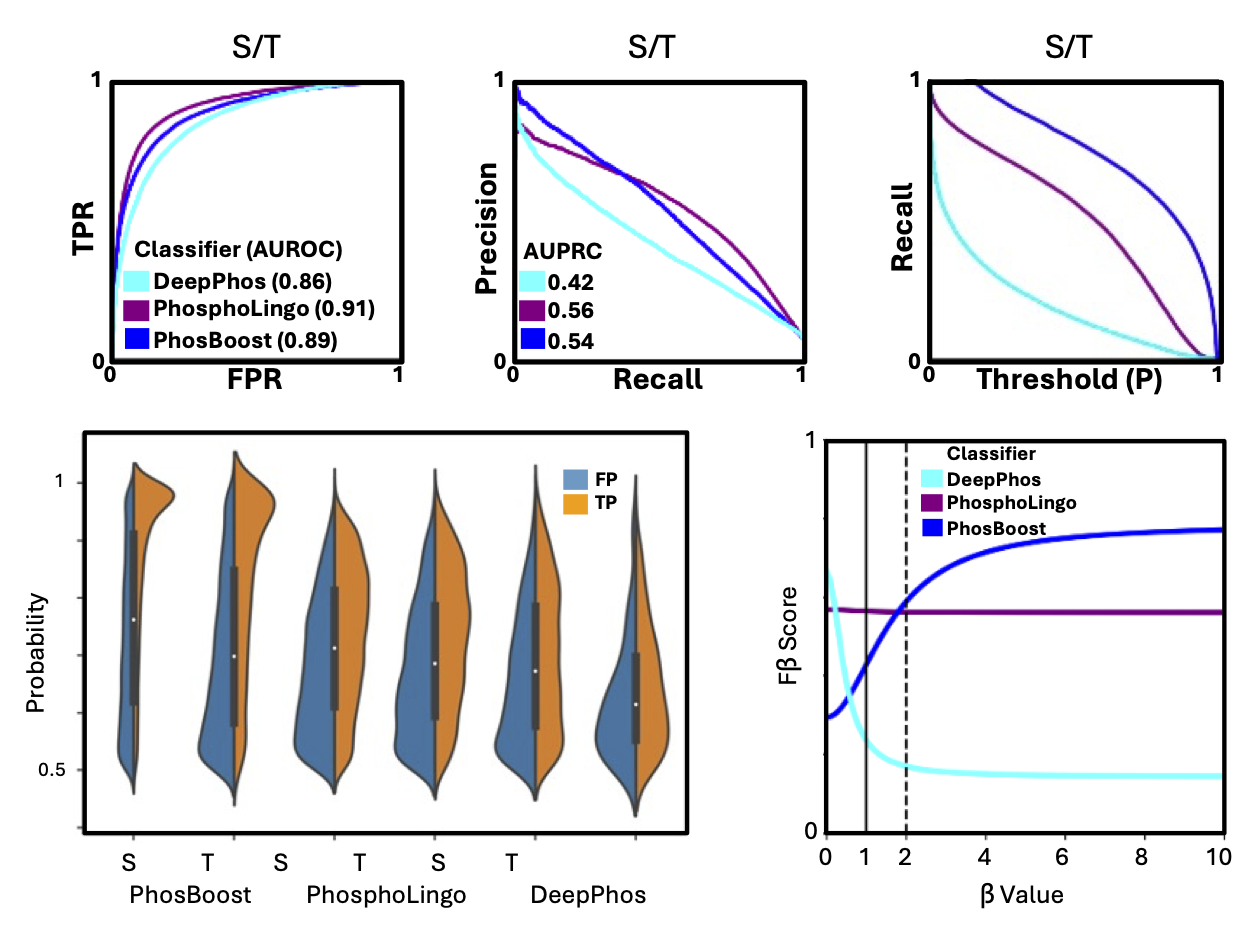
PhosBoost
A Colab notebook I prepared for predicting protein phosphorylation sites for a user-provided protein, based on our PhosBoost manuscript. The Colab notebook runs fast on a CPU node, so it should also work with a free account. Additional details can be found in the paper, GitHub repository, and the Colab notebook.
AllMetal3D
##
A minimal Colab notebook for predicting metal and water binding sites in proteins. Works with CUDA out of the box when running on a GPU node.
AlphaFill-Relax
A Colab notebook I prepared for running a brief 20ps Molecular Dynamics (MD) equilibration simulation step using OpenMM engine and AMBER force field. My very preliminary results show that it can improve the positioning of metal cofactors in terpene synthase active sites. Additional information can be found in the bioinformatics/proteins page and inside the notebook.

Relax Amber Batch
A Colab notebook I prepared for running the amber relaxation step recommended for predicted protein structures. This notebook is based completely on the relax_amber notebook that can be found at the ColabFold repository. The only addition offered here is the ability to batch-apply the PDB relaxation step to all PDB files in a given Google Drive folder.
